| x | x | |||||
 |
||||||
| BACTERIOLOGY | IMMUNOLOGY | MYCOLOGY | PARASITOLOGY | VIROLOGY | ||
|
|
|
|||||
| En Español | ||||||
|
Let us know what you think FEEDBACK |
||||||
| SEARCH | ||||||
|
|
||||||
|
|
||||||

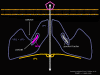
Figure 1a RNA virus replication strategies 
Figure 1b RNA viruses without a DNA phase 
Figure 1c RNA viruses with a DNA phase: retroviruses |
Most RNA viruses have single strand genomes, rather than a double helical RNA (in contrast, DNA viruses contain double helical DNA, as is found in eukaryotic cell DNA). This is because DNA is more easily unwound during transcription whereas double stranded RNA is very stable (its melting temperature is ten degrees higher than a DNA of similar composition). Moreover, unwinding of RNA is not a host cell mechanism, in contrast to DNA unwinding that takes place whenever cellular DNA is transcribed to DNA or to mRNA. There are two basic types of RNA virus, those that copy their RNA to new RNA without a DNA intermediate and those that use a DNA intermediate (figure 1a and b). The former include positive strand RNA viruses (e.g. picornaviruses such as polio and rhino viruses), negative strand RNA viruses (e.g. influenza) and double strand RNA viruses (e.g. rotaviruses). The RNA viruses that have a DNA intermediate are the retroviruses (figure 1 a and c).
Animal RNA viruses encode a polymerase Since copying RNA to RNA is not a host cell function, RNA viruses that do not have a DNA stage must code for an RNA-dependent RNA polymerase (this is also called a replicase). Such viruses typically go through their entire life cycle in the cytoplasm. Retroviruses are positive strand RNA viruses and must encode an RNA-dependent DNA polymerase (reverse transcriptase) to make the DNA provirus which then is transcribed to genomic RNA by a host enzyme, RNA polymerase II. Since RNA polymerase II makes mRNA in the nucleus of the host cell, the life cycle of retroviruses takes place mostly in the nucleus. There is one exception to the notion that all animal RNA viruses encode their own polymerase and this is hepatitis delta agent which is not a true virus. It may use a host enzyme that normally copies DNA to RNA to copy its RNA to RNA.
|
|||||
|
|
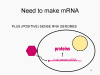
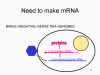
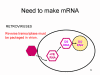
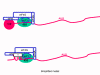
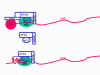
Viral Messenger RNA An RNA virus needs to make an RNA that can serve as a messenger RNA for protein synthesis in a host cell. In the case of the positive strand RNA viruses (whose genome, by definition, is the same sense as mRNA), the genomic RNA can serve as the message (figure 2). The virus capsid serves as the delivery vehicle to the cytoplasm. Since the genome is RNA and copied by an RNA polymerase, there is no need for a typical promoter (TATA box, CAT box etc) upstream of the protein encoding genes (as found in DNA viruses or retroviruses); moreover, positive strand viruses do not need to make new proteins before making mRNA as their RNA can serve directly as a message. In contrast, cells infected by a negative strand viruses must contain a protein (the polymerase) before positive sense mRNA can be made and so the virus must carry the polymerase inside the capsid and deliver this protein to the infected cell (figure 3). In addition, the sense mRNA that is copied from the incoming negative sense genomic RNA must have the normal modifications of the cell’s mRNA (methyl cap, poly A tail) in order to be translated by the host cell. This means that modifying enzymes (methylase, GMP transferase, poly A polymerase) must also be packaged in the incoming virus. Although the host cell could provide these enzymes, they are located in the nucleus (where mRNA is made) and, as we have noted above, most RNA viruses are cytoplasmic throughout their life cycle. In the case of retroviruses, copying of RNA to DNA is necessary before protein can be made and so the polymerase (reverse transcriptase) must also be packaged in the virus particle (figure 4). These RNA viruses, however, replicate their genome in the host cell's nucleus and so can use host cell RNA modification enzymes.
The monocistronic RNA problem Eukaryotic cells have monocistronic mRNAs, in contrast to bacteria that can have polycistronic mRNAs. A monocistronic mRNA codes for one protein since the ribosome finds a translation initiation site near the 5’ end of the mRNA and translates the protein until it come to a stop codon. Internal initiation of protein synthesis within the mRNA does not occur (except in rare situations). A single RNA can code for more than one protein in eukaryotes but this is done by splicing out parts of the original transcript to make another mRNA that serves as a monocistronic message. Splicing enzymes that carry out this process are found in the nucleus (since this is where mRNAs are made). Since RNA viruses are normally cytoplasmic, they cannot take advantage of splicing enzymes. Thus, RNA viruses that have only one mRNA ought only to be able to make one large protein - but they have developed a number of tricks to overcome this (figure 5) and do, indeed, make more than one protein. Some can take advantage of the alternative splicing enzymes of the host cells (and therefore must have a nuclear stage). Others make a single large protein which has a protease activity; this cuts up the large precursor to a series of smaller proteins. Others, such as the picornaviruses, have found ways to make a single mRNA function in a polycistronic fashion even though they are in a eukaryotic cell. How they do this will be addressed later.
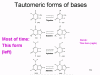
The proof reading problem When we copy our DNA genome, we use a proof reading DNA polymerase that corrects mistakes in the new copy of the DNA. Most errors occur because the normal forms of the bases that make up the code in DNA can exist in rare tautomeric forms (figure 6). These rare forms exist for very short periods of time but if a base is in its tautomeric form at the time that it is copied, it will be read as the wrong base during DNA synthesis. Thus, normally A pairs with T but the rare tautomer of T will base pair with G and be copied accordingly. RNA polymerases do not normally have proof reading and so these mutations will not be corrected and will be perpetuated in future generations. The mutation rate of RNA polymerase II is about 1 in 10,000 and so, to keep the mutation load of the RNA viruses down, they must have small genomes; usually, RNA viruses have genomes that are 10,000 nucleotides or smaller (compared, for example, to herpes viruses that have genomes of about 150,000 nucleotides).
Why do all these mutations not make RNA viruses unstable? The answer is that they do in many cases. Thus, HIV (which has a genome size of just fewer than 10,000 nucleotides) accumulates many mutations which result in population polymorphism and allow the virus to become resistant to anti-viral drugs. However, many RNA viruses (e.g. polio virus) are very stable over thousands of years despite their high rate of mutation. We can best explain this with the concept of fitness in a Mullerian landscape. According to this concept, a certain virus strain may be well adapted to replicate in a certain host (e.g. polio in humans). There may be other forms of the virus that could be equally or more successful but to mutate to another form the virus would have to go through a form that is less well adapted. Thus, when the well adapted virus is mutated, the mutant viruses cannot compete and are diluted out. In other words, well adapted viruses have a peak of fitness in their niche and are unable to cross the valley of poorer adaptation (figure 7). This is rather like the squirrels on the north and south rims of the Grand Canyon which have diverged because they cannot adapt to the harsh conditions on the canyon floor and therefore cannot interbreed.
Consequences of a small genome size A small genome restricts the number of proteins that can be encoded (figure 8). There needs to be a capsid protein to protect the RNA and a polymerase which, as we have seen, must be made by the virus. Also necessary will be an attachment protein so that the virus can infect a new cell. Thus, there will be a limited number of individual proteins although RNA viruses can be quite large by using multiple copies of the various proteins.
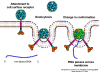
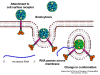
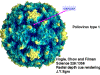
Life cycle Polio (figure 9) and rhinoviruses are picornaviruses. They are small, icosahedral, positive sense RNA viruses that do not have an envelope (lipid bilayer). The picornaviruses also include hepatitis A virus and enteroviruses. The entire life cycle of these viruses occurs in the cytoplasm of the host cell. Since the RNA is the same sense as mRNA, the RNA alone is infectious, though to a very much lesser extent than the complete virus particle. This is because the virus has proteins that attach to the cell surface. Picornaviruses attach to the host cell via a specific receptor, as do virtually all other viruses and are internalized by the cell (figure 10). In the case of polio and rhino viruses, this is a protein of unknown function (in normal cells) known as CD155, the poliovirus receptor or PVR. We do have some clues about CD155's normal function in that it binds to a protein called vitronectin and so may be an adhesion protein involved in the binding of a cell to its extracellular matrix. A majority of rhinoviruses can also bind to another receptor which is an adhesion protein called CD54 or ICAM-1 (intercellular adhesion molecule-1); the use of these receptors restricts the cells that the viruses can infect, leading to their characteristic tropism; for example, CD155 is expressed on several cell types including in the gut and also at the terminal of an axon at the neuromuscular junction, that is at the presynaptic membrane. CD155 and ICAM-1 are glycoproteins that are anchored in the membrane which they span once (figure 11D). Their extracellular regions, to which the picornavirus binds, have three and five immunoglobulin-like domains, respectively, with immunoglobulin-like folds. In each case, the virus binds to the N-terminal domain. ICAM-1 is longer and protrudes deep into the canyon (figure 11C,D) while CD155 is shorter lies along the canyon (figure 11E). If you look at the structure of the nucleocapsid you will see twelve pentons, one at each vertex of the virus. Each penton is in contact with five other proteins that surround it. Each face of the virus has a protein complex that looks like a triskelion (a three legged structure). This is in contact with six other proteins (three pentons and three triskelions) and is sometimes called a hexon, though it does not look like a hexon in the way that the penton looks like a penton. Since the icosahedron has twenty faces, there are 20 triskelions. Between the pentons and the triskelions is a canyon (figure 11A,B); the receptor on the cell surface (ICAM-1 or CD155) binds in the canyon (figure 11E) and precipitates a conformational change in some of the viral proteins. The interaction of cell and viral protein is not very stable. In the case of rhinoviruses, entry into the cytoplasm seems to follow endocytosis which might suggest that an acid pH is required for the conformational change. Let’s look in more detail at what happens when a rhinovirus binds to the cell via ICAM-1. The RNA will enter the cell though the membrane via the center of the penton after one of the viral proteins embeds in the cell’s membrane. If we take a cross section through the penton (figure 12) (which has five fold symmetry when we look down on it), we can see that VP1 molecules form the central part of the penton and surround a closed pore. The canyon is located where the VP1 apposes VP2/3. But the canyon floor is not flat. There is a narrow hole in the floor under which is a bigger space, known as the pocket. Normally, this pocket contains “pocket factor”. This has never been isolated and characterized but it appears as a long fatty acid-like molecule in crystallographic structural studies. Below the external proteins, VP1, VP2 and VP3, is another virus protein that cannot be seen from the outside (as in figure 12) and this is VP4. Figure 13 shows the backbone of the proteins of the penton with a more general structure in the inset. Figure 14 shows how we think that the ICAM-1 molecule fits into the VP1/2/3 complex. Note that it fits into the canyon but does not penetrate the pocket. We see this again in figure 15 with the backbones of the proteins now indicated. One class of drugs that has been used to treat picornavirus infections is the WIN drugs. They stop the conformational change that occurs when the virus and ICAM-1 interact and in figure 16 you can see what happens. The WIN drug displaces “pocket factor” and fits snuggly into the pocket (figure 17) and is almost completely buried as can be seen when we look at the penton in a space-filling image (figure 18). As we noted above, one protein, VP4, is underneath the external VP1/2/3 protein. VP4 has a fatty acid (myristic acid) attached to it (figure 19) which makes one end very hydrophobic. In the space-filling model (figure 20), we can see that the fatty acid protrudes into the pore to form a plug at the middle of the penton. VP3 also forms part of the plug (figure 20). When ICAM-1 binds in the canyon, there is a conformational change and VP1 tips back away from the center of the penton (figure 21). This squeezes the pocket and it is possible that binding displaces the pocket factor. Now you can see why the WIN drugs work. They bind in the pocket more strongly than pocket factor and are not displaced on ICAM-1 binding. Thus, the bend to open the pore does not occur (figure 21b). When VP1 tips back (figure 21b,c), the pore opens as VP3 moves out of the way. This allows VP4 to move through the pore (channel). The amino terminus of VP4 has a myristic acid (fatty acid) and this in some way moves through the pore. Remember that the pore has five-fold symmetry and there are really five of these VP4 proteins being forced up the channel. The myristic acids embed in the lipid bilayer of the cell’s membrane to form a pore. The formation of the pore is also helped by the fact that the VP1 amino terminus is an amphipathic alpha helix which flips out on ICAM-1 binding. As might be expected from the structure of the
VP1-4 tetramer, VP4 should not be bound by anti-VP4 antibodies on the
outside of the virus. If we leave the virus for a long time in solution,
VP4 does get labeled as the virus “breaths”. When a WIN drug is bound
in the pocket such breathing stops showing that the tighter binding of
the drug in the pocket stabilizes the structure of the virus. As might
be expected, if we add a part of the ICAM-1 molecule that binds in the
canyon, there is a shape change and the RNA comes out of the pore and
the virus is no longer infectious. The penton is not only the site of
the RNA exit into the cell that is being infected, it is also the entry
site for the RNA into the nucleocapsid. |
|||||
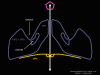
Figure 12 Arrangement of the proteins of the rhinovirus penton
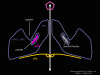 Figure 16 WIN drugs displace the pocket factor
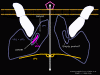 Figure 21b. When ICAM-1 binds pocket factor is probably displaced and the VP1,2,3 complex tips to open the pore. When a WIN drug has replaced pocket factor, this conformational change does not occur
|

Figure 13 Backbone structure of the proteins of the penton

Figure 21c Binding of ICAM-1 results in opening of the pore, the embedding of parts of VP1 and VP4 (which may also pass through the pore) in the membrane of the cell to which the virus has bound. This forms a pore in the cell membrane. The RNA can pass through the pore of the virus penton and into the host cell
|
|||||

Figure 22 VpG is at the 5' end of the viral genomic sense (+ve) RNA but is lost before translation
|
Once the RNA has entered the cell it can be translated. At the 5’ end of the RNA there is a small viral protein called VPg (figure 22). This is removed from the RNA when the cell is infected (figure 22) and since the RNA is positive sense, it can now be used as mRNA. Normally, when a cellular mRNA is translated, the small ribosomal subunit recognizes the cap structure at the 5’ end and then the ribosome assembles after the binding of the large subunit. But there is no methylated cap on the picornavirus RNA since capping occurs in the nucleus (as a result of cellular enzymes) and the entire life cycle of a picornavirus takes place in the cytoplasm. Nevertheless, the host protein synthesis machinery does translate the viral RNA. Figure 23 shows the normal initiation complex that forms on a capped mRNA. In the picornavirus RNA there is a region of secondary structure called an internal ribosome entry site (IRES) (figure 24). This is recognized by the complex of proteins that normally recognizes the cap. There appears to be an additional host cell factor that is necessary for the recognition of an IRES but not for a normal mRNA methyl cap. It is possible that the availability of this host factor determines in part the tropism of picornaviruses for certain cells. Picornaviruses also have a protease activity that can cut one of the proteins of the initiation complex, eIF4G (figure 25), and this seriously affects the cell’s ability to translates normal capped messages but does not affect translation from the IRES; thus the virus can suppress host cell translation while leaving the translation of its own RNA unaffected. The fact that there is only one IRES means that there is only one primary translation product; that is there can only be one large protein made. This protein will eventually be cut up to make several proteins in the mature virus and thus we call this primary product a polycistronic protein since it is encodes by more than one gene (cistron). The proteases that cut up the original polyprotein are encoded in the virus genome and the proteolytic process is ordered as shown in figure 26.
|
|||||
 Figure 24 In the picornavirus RNA there is a region of secondary structure called an internal ribosome entry site (IRES)
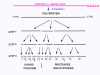 Figure 26 The proteases that cut up the original polyprotein are encoded in the virus genome and the proteolytic process is ordered |
||||||
 A
A
|
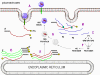
This is quite simple compared to some other RNA viruses. Since picornaviruses spend all of their time in the cytoplasm, they must encode a polymerase (replicase) that is made from the sense strand of the infecting virus. The polymerase copies the sense strand to anti-sense which is then copied back to the sense strand that is packaged into the virus (figure 27). RNA replication seems to occur on the cytoplasmic surface of membrane vesicles to which the RNA polymerase binds. These appear to come from the endoplasmic reticulum, as do vesicles in the uninfected cells that transport secreted and membrane proteins to the Golgi body. However, when the cell is infected by the picornavirus, the vesicles do not fuse with the cis face of the Golgi body as the transport vesicles do (figure 28). These vesicles have specific targeting proteins on their cytoplasmic surfaces (called COP proteins) and it is possible that they are involved in the response to the viral infection. It should be remembered that picornaviruses do not have a lipid envelope and do not have a surface glycoprotein. Therefore, the production of virus is not inhibited by compromising Golgi Body function, as would be the case with an enveloped virus. We do not know why there is this membrane association of RNA replication of picornaviruses but it may concentrate various substrates in the vicinity of the polymerase (remember that in bacteria, DNA replication is membrane-associated). The RNA of picornaviruses is polyadenylated at the 3’ end, as are cellular messenger RNAs but this polyadenylation occurs in a different way. In host cell mRNA synthesis, the poly A sequence is not coded in the DNA copy of the gene but is added by an enzyme called poly A polymerase using ATP as a substrate. In the case of picornavirus RNA, however, the poly A sequence in the sense strand is copied to a poly U sequence at the 5’ end of the negative strand. This is copied back to 3’ poly A, again by the replicase.
|
|||||
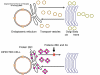
Figure 28 When the cell is infected by a picornavirus, the endoplasmic reticulum to Golgi body transport vesicles do not fuse with the cis face of the Golgi body |
ASSEMBLY As has been noted above, picornavirus proteins are polycistronic because there is only one internal ribosome binding site. This means that the virus can be assembled as the various proteins are cleaved from the polyprotein. The polyprotein is first cleaved to three proteins (P1, P2, P3). P1 is then cleaved into three proteins (VP0, VP1, VP3) that make up the subunit of the virus coat. This is done by virus-specified proteases that are part of the polyprotein and are catalytic as part of the single primary translation product. VP0, VP1 and VP3 assemble into the 5S structural subunit (protomer). VP0is only cleaved to VP2 and VP4 when the virus has assembled. Five of these protomers assemble into a 14S pentamer and twelve pentamers form the procapsid. RNA is encapsulated into the procapsid to form a provirion. At this stage VP0 is cleaved to VP2 and VP4 and the virion is a mature, infectious virus particle. VP0 is only cleaved if the RNA is encapsidated. The other two parts of the primary translation product (P2 and P3) are cleaved to form a number of non-structural proteins (i.e. proteins that are not found in the mature virus particle but which are used during replication in the infected cell). These include the replicase and proteins that alter host cell metabolism. VPg, the protein that is found at the end of each of the positive sense genomic RNA molecules is formed from part of P3. |
|||||
|
RELEASE Many picornavirus particles accumulate in the cytoplasm and the cell dies resulting in cellular lysis. |
||||||
|
Thus the entire life cycle of polio virus occurs in the cytoplasm
|
||||||
|
||||||